Expected Results
ΔGqq ∆Gqq bar graphs analysis
The TKSA-MC method indicates the residues which contribute to destabilizing the protein native state. The algorithm calculates the protein electrostatic energy taking into account the contribution of each residue with polar charged side chain. Figure 1 presents an example of ΔGqq bar graph for ubiquitin protein 1.
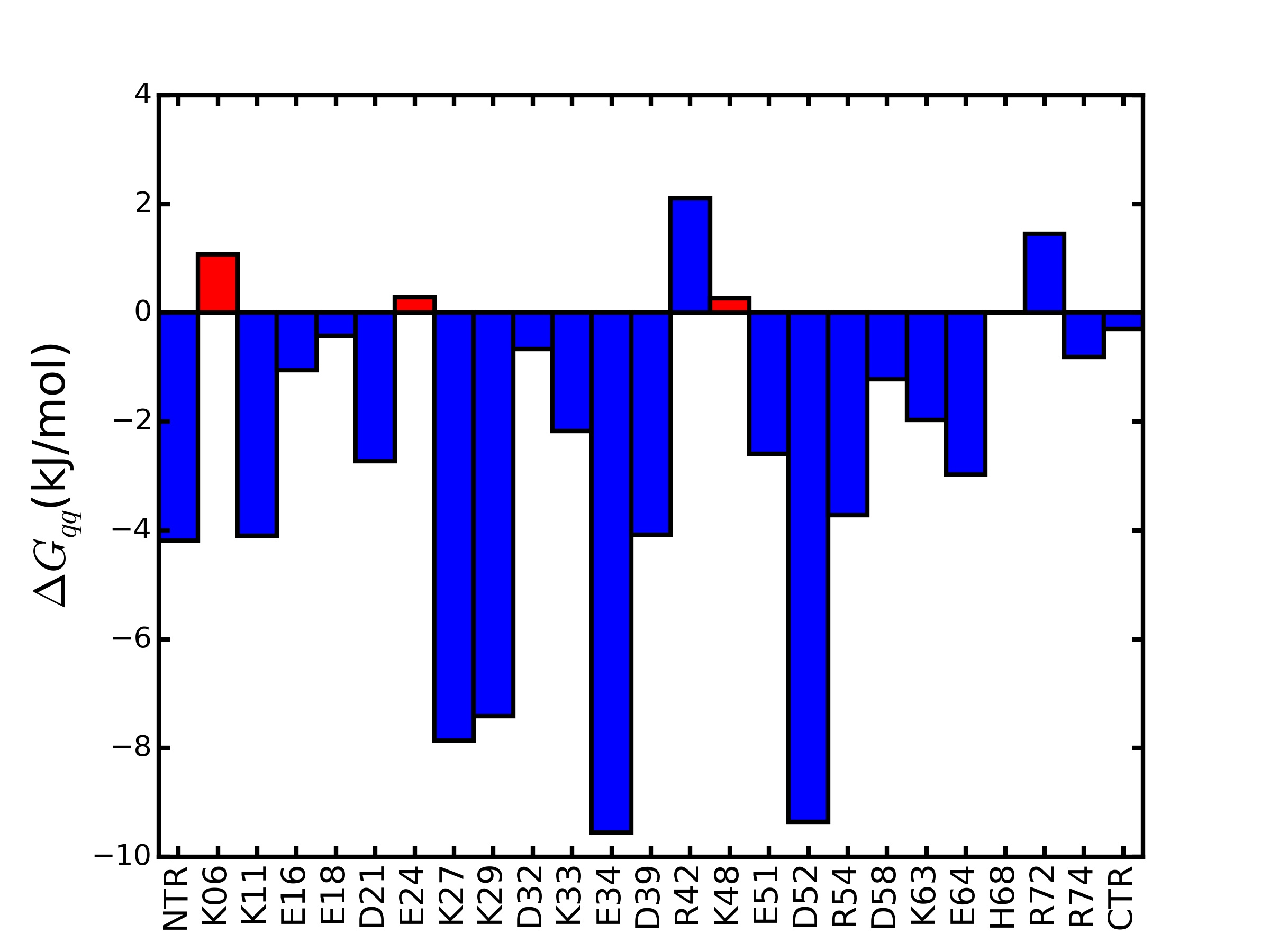
The bars indicate the charge-charge energy contribution of each ionizable residue to the protein native state stability. The red bars indicate the candidate residues to be mutated to increase the protein thermostability and the selection criteria follows the methodology introduce by Ibarra-Molero et al.2. It was indicated the residue which present unfavorable energy values ΔGqq ≥ 0 and are exposed to solvent with SASA ≥ 50%. In ubiquitin example the highlighted residues are K06, E24 and K48 and this information is also printed in result screen page with its respective ΔGqq and SASA values. All the used data to plot the ΔGqq bar graphs figure is available for download in the output files. The total energy contribution to protein native state stability is given as a sum of the ΔGqq for each ionizable group n in the protein, named as ΔGelec. The ΔGelec for the ubiquitin protein in pH 5.5 is -61.99kJ/mol.
ΔGelec and pH dependence
The total energy contribution ΔGelec is a pH dependent measure. The ionization degree of each polar residue side chain and its ΔGqq vary with the pH input value. Figure 2 presents an example of ΔGelec as a function of pH for ubiquitin protein.
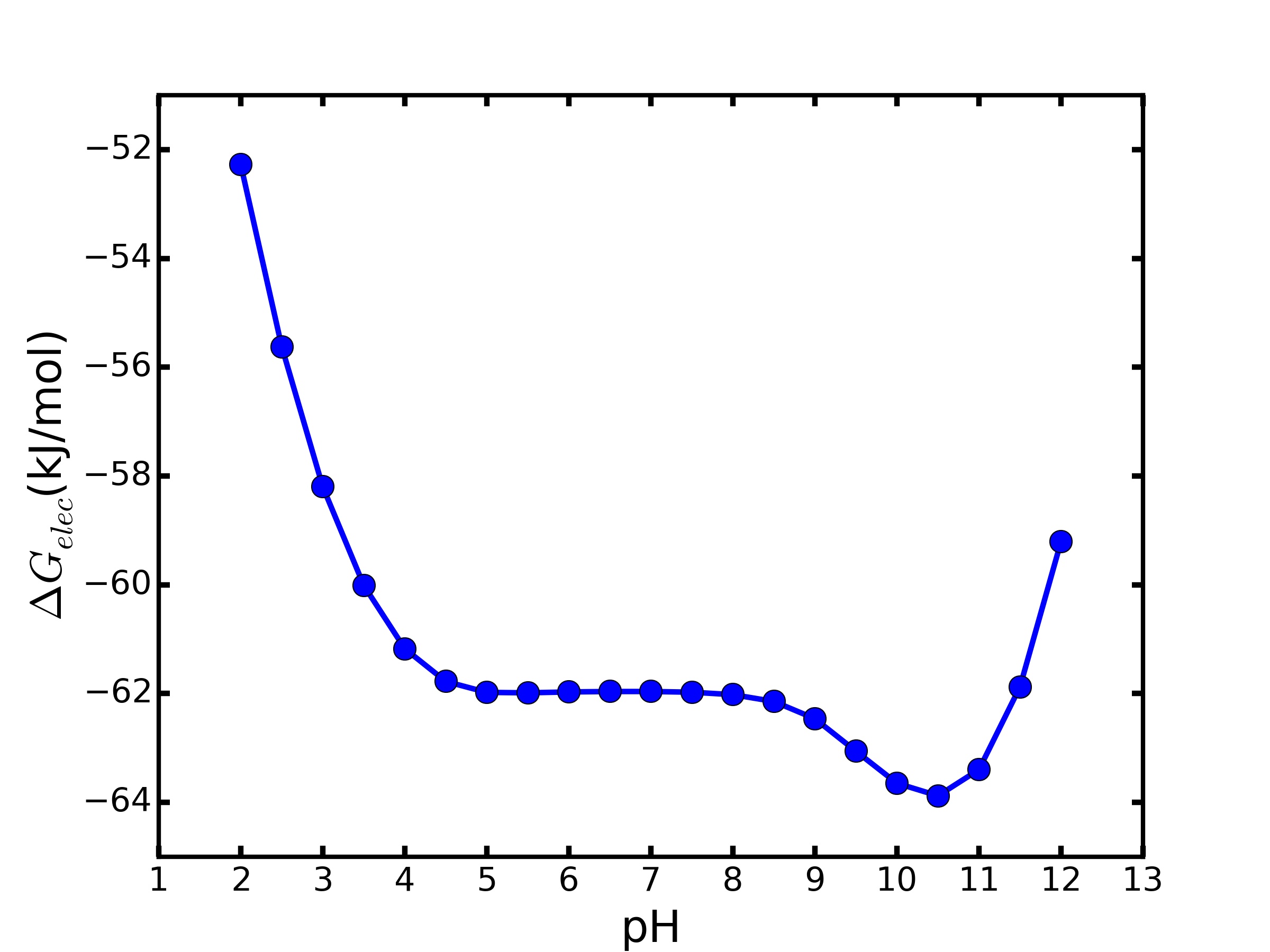
The ΔGelec curve indicates the pH values which the electrostatic energy contribution of each residue to stabilizes the protein native state. The pH range used in Figure 2 result was from 2 to 12 in step of 0.5 in a total of 21 TKSA-MC runs. All the used data to plot the ΔGelec curve and the respective pH bar graphs figures are available for download in the output files.
References
(1) Strickler, S. S.; Gribenko, A. V.; Gribenko, A. V.; Keiffer, T. R.; Tomlinson, J.; Reihle, T.; Loladze, V. V.; Makhatadze, G. I. Biochemistry 2006, 45, 2761–2766.
(2) Ibarra-Molero, B.; Loladze, V. V.; Makhatadze, G. I.; Sanchez-Ruiz, J. M. Biochemistry 1999, 38, 8138–8149.